Research
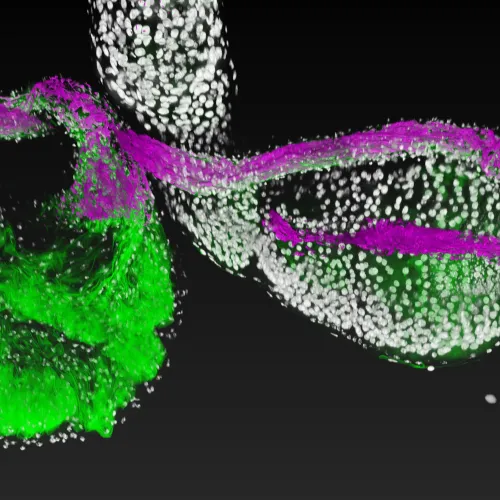
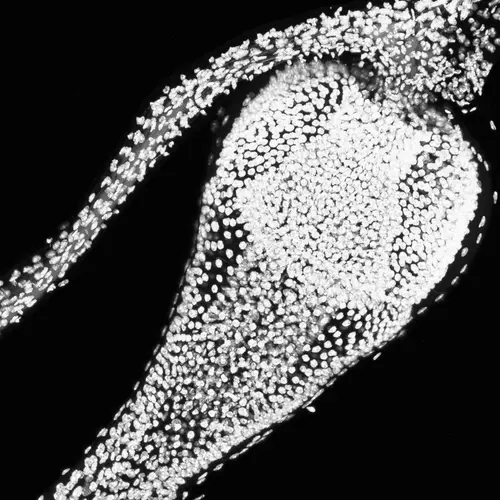
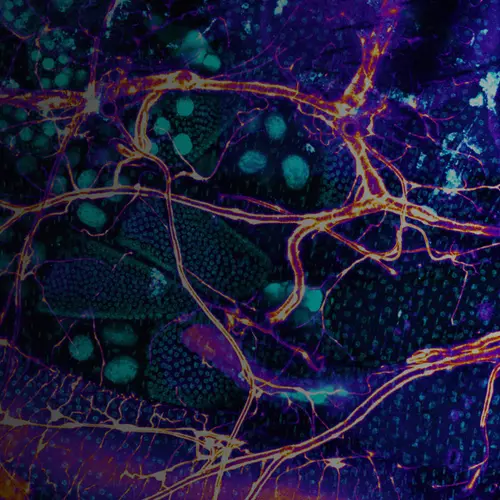
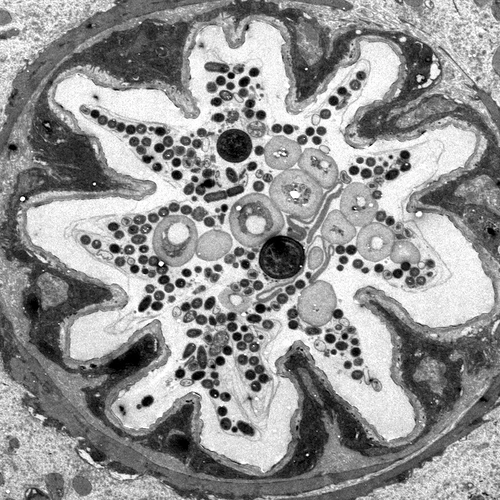
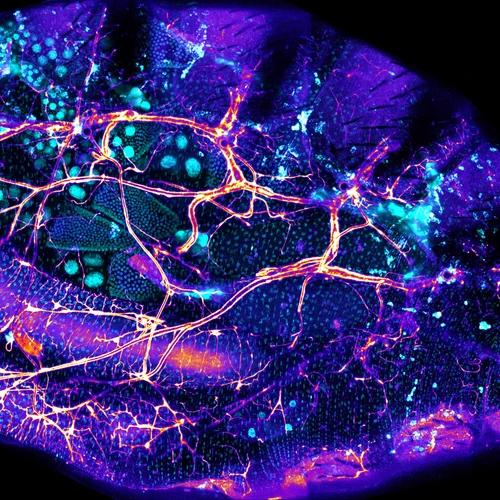
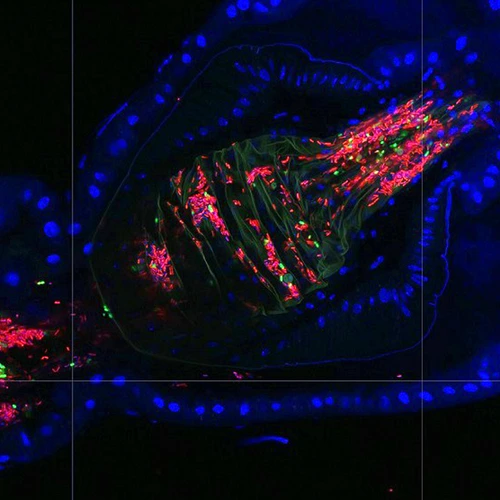
The intestines of animals are typically colonized by a complex, relatively stable microbiota that influences health and fitness, but the underlying mechanisms of colonization remain poorly understood. As a typical animal, the fruit fly, Drosophila melanogaster, is associated with a consistent set of commensal bacterial species, yet the reason for this consistency is unknown. We use gnotobiotic flies, genetics, microscopy, and microbiology techniques to examine the development and maintenance of a defined region in the Drosophila foregut that selects and maintains a multispecies community of bacteria with strain-level specificity.
How is exquisite regulation achieved? What does the host control? How do bacterial interactions affect the community composition? How do these relationships evolve?
Current Topics
We recently identified a physical niche structure in the fly gut that houses specific bacterial symbionts. We examine the fly genetics of gut symbiosis including the construction of the microbial niche.
Using both gnotobiotic flies and high throughput in vitro growth assays, we study the microbial interactions that shape the fly gut microbiota.
Using gnotobiotic flies and a battery of physiological and fitness assays, we study the effects of gut microbial community ecology on host cell physiology.
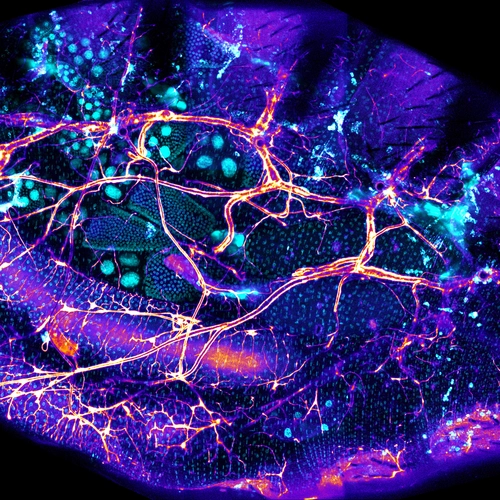
We have developed single-fly feeding and live fly microscopy techniques to measure microbial populations in the gut. These approaches allow us to visualize the process of gut colonization in real time to understand how the fly gut selects the correct bacterial species for colonization.
Recent Publications
Abstract: The gut is continuously invaded by diverse bacteria from the diet and the environment, yet microbiome composition is relatively stable over time for host species ranging from mammals to insects, suggesting host-specific factors may selectively maintain key species of bacteria. To investigate host specificity, we used gnotobiotic Drosophila, microbial pulse-chase protocols, and microscopy to investigate the stability of different strains of bacteria in the fly gut. We show that a host-constructed physical niche in the foregut selectively binds bacteria with strain-level specificity, stabilizing their colonization. Primary colonizers saturate the niche and exclude secondary colonizers of the same strain, but initial colonization by Lactobacillus species physically remodels the niche through production of a glycan-rich secretion to favor secondary colonization by unrelated commensals in the Acetobacter genus. Our results provide a mechanistic framework for understanding the establishment and stability of a multi-species intestinal microbiome.
LinkAbstract: Non-mammalian model organisms have been essential for our understanding of the mechanisms that control development, disease, and physiology, but they are underutilized in pharmacological and toxicological phenotypic screening assays due to their low throughput in comparison with cell-based screens. To increase the utility of using Drosophila melanogaster in screening, we designed the Whole Animal Feeding FLat (WAFFL), a novel, flexible, and complete system for feeding, monitoring, and assaying flies in a high-throughput format. Our 3D printed system is compatible with inexpensive and readily available, commercial 96-well plate consumables and equipment. Experimenters can change the diet at will during the experiment and video record for behavior analysis, enabling precise dosing, measurement of feeding, and analysis of behavior in a 96-well plate format.
LinkAbstract: The intestines of animals are colonized by commensal microbes, which impact host development, health, and behavior. Precise quantification of colonization is essential for studying the complex interactions between host and microbe both to validate the microbial composition and study its effects. Drosophila melanogaster, which has a low native microbial diversity and is economical to rear with defined microbial composition, has emerged as a model organism for studying the gut microbiome. Analyzing the microbiome of an individual organism requires identification of which microbial species are present and quantification of their absolute abundance. This article presents a method for the analysis of a large number of individual fly microbiomes. The flies are prepared in 96-well plates, enabling the handling of a large number of samples at once. Microbial abundance is quantified by plating up to 96 whole fly homogenates on a single agar plate in an array of spots and then counting the colony forming units (CFUs) that grow in each spot. This plating system is paired with an automated CFU quantification platform, which incorporates photography of the plates, differentiation of fluorescent colonies, and automated counting of the colonies using an ImageJ plugin. Advantages are that (i) this method is sensitive enough to detect differences between treatments, (ii) the spot plating method is as accurate as traditional plating methods, and (iii) the automated counting process is accurate and faster than manual counting. The workflow presented here enables high-throughput quantification of CFUs in a large number of replicates and can be applied to other microbiology study systems including in vitro and other small animal models.
LinkAbstract: Observational studies reveal substantial variability in microbiome composition across individuals. Targeted studies in gnotobiotic animals underscore this variability by showing that some bacterial strains colonize deterministically, while others colonize stochastically. While some of this variability can be explained by external factors like environmental, dietary, and genetic differences between individuals, in this paper we show that for the model organism Drosophila melanogaster, interactions between bacteria can affect the microbiome assembly process, contributing to a baseline level of microbiome variability even among isogenic organisms that are identically reared, housed, and fed. In germ-free flies fed known combinations of bacterial species, we find that some species colonize more frequently than others even when fed at the same high concentration. We develop an ecological technique that infers the presence of interactions between bacterial species based on their colonization odds in different contexts, requiring only presence/absence data from two-species experiments. We use a progressive sequence of probabilistic models, in which the colonization of each bacterial species is treated as an independent stochastic process, to reproduce the empirical distributions of colonization outcomes across experiments. We find that incorporating context-dependent interactions substantially improves the performance of the models. Stochastic, context-dependent microbiome assembly underlies clinical therapies like fecal microbiota transplantation and probiotic administration and should inform the design of synthetic fecal transplants and dosing regimes.
LinkA longstanding goal of biology is to identify the key genes and species that critically impact evolution, ecology, and health. Yet biological interactions between genes (1, 2), species (3–6), and different environmental contexts (7–9) change the individual effects due to non-additive interactions, known as epistasis. In the fitness landscape concept, each gene/organism/environment is modeled as a separate biological dimension (10), yielding a high dimensional landscape, with epistasis adding local peaks and valleys to the landscape. Massive efforts have defined dense epistasis networks on a genome-wide scale (2), but these have mostly been limited to pairwise, or two-dimensional, interactions (11). Here we develop a new mathematical formalism that allows us to quantify interactions at high dimensionality in genetics and the microbiome. We then generate and also reanalyze combinatorically complete datasets (two genetic, two microbiome). In higher dimensions, we find that key genes (e.g. pykF) and species (e.g. Lactobacillus plantarum) distort the fitness landscape, changing the interactions for many other genes/species. These distortions can fracture a “smooth” landscape with one optimal fitness peak into a landscape with many local optima, regulating evolutionary or ecological diversification (12), which may explain how a probiotic bacterium can stabilize the gut microbiome.
LinkTimeline
All Publications
- A chemically-defined growth medium to support Lactobacillus-Acetobacter sp. community analysis. K Aumiller, R Scheffler, ET Stevens, ZT Güvener, E Tung, AB Grimaldo, ... PLoS One 18 (10), e0292585; 2023
- Gut microbiome dysbiosis is associated with host genetics in the Norwegian Lundehund. C Melis, AM Billing, PA Wold, WB Ludington. Frontiers in Microbiology 14, 1209158; 2023
- Expanding evolutionary theories of ageing to better account for symbioses and interactions throughout the Web of Life. E Bapteste, P Huneman, L Keller, J Teulière, P Lopez, EC Teeling, .... Ageing Research Reviews, 101982; 2023
- Signal in the noise: temporal variation in exponentially growing populations. EW Jones, J Derrick, RM Nisbet, W Ludington, DA Sivak. arXiv preprint arXiv:2304.11474; 2023
- Pulsed, continuous or somewhere in between? Resource dynamics matter in the optimisation of microbial communities. AD Letten, WB Ludington. The ISME Journal 17 (4), 641-644; 2023
- A symbiotic physical niche in Drosophila melanogaster regulates stable association of a multi-species gut microbiota. R Dodge, EW Jones, H Zhu, B Obadia, DJ Martinez, C Wang, .... Nature Communications 14 (1), 1557; 2023
- Whole Animal Feeding FLat (WAFFL): a complete and comprehensive validation of a novel, high-throughput fly experimentation system. MDLA Jaime, GH Salem, DJ Martinez, S Karott, A Flores, CD Palmer, .... G3: Genes, Genomes, Genetics 13 (3), jkad012; 2023
- From worms to humans: Understanding intestinal lipid metabolism via model organisms. DW Kozan, JT Derrick, WB Ludington, SA Farber. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 159290; 2023
- Fast Colony Forming Unit Counting in 96-Well Plate Format Applied to the Drosophila Microbiome. R Dodge, WB Ludington. Journal of Visualized Experiments, doi:10.3791/64298; 2023
- Nutrient encryption and the diversity of cobamides, siderophores, and glycans. ME Taga, WB Ludington. Trends in Microbiology; 2022
- Higher-order microbiome interactions and how to find them. WB Ludington. Trends in Microbiology; 2022
- Microbiome-by-ethanol interactions impact Drosophila melanogaster fitness, physiology, and behavior. JA Chandler, LV Innocent, DJ Martinez, IL Huang, JL Yang, MB Eisen, .... iScience 25 (4); 2022
- Stochastic microbiome assembly depends on context. EW Jones, JM Carlson, DA Sivak, WB Ludington. Proceedings of the National Academy of Sciences 119 (7), e2115877119; 2022
- Genetic rescue of the highly inbred Norwegian Lundehund. C Melis, C Pertoldi, WB Ludington, C Beuchat, G Qvigstad, AV Stronen. Genes 13 (1), 163; 2022
- From a parts list to assembly instructions and an operating manual: how small host models can re-write microbiome theory. NM Vega, WB Ludington. Current Opinion in Microbiology 64, 146-151 1; 2021
- High dimensional geometry of fitness landscapes identifies master regulators of evolution and the microbiome. H Eble, M Joswig, L Lamberti, WB Ludington. bioRxiv, 2021.09. 11.459926; 2021
- Master regulators of evolution and the microbiome in higher dimensions. H Eble, M Joswig, L Lamberti, W Ludington. arXiv preprint arXiv:2009.12277; 2020
- Drosophila as a model for the gut microbiome. WB Ludington, WW Ja, PLoS Pathogens 16 (4), e1008398; 2020
- Testing the role of intraflagellar transport in flagellar length control using length-altering mutants of Chlamydomonas. K Wemmer, W Ludington, WF Marshall. Philosophical Transactions of the Royal Society B 375 (1792), 20190159; 2020
- Bacterial interspecies interactions modulate pH-mediated antibiotic tolerance. A Aranda-Díaz, B Obadia, R Dodge, T Thomsen, ZF Hallberg, ZT Güvener, .... eLife 9, e51493; 2020
- Bellymount enables longitudinal, intravital imaging of abdominal organs and the gut microbiota in adult Drosophila. LAJ Koyama, A Aranda-Díaz, YH Su, S Balachandra, JL Martin, .... PLoS Biology 18 (1), e3000567; 2020
- Cluster partitions and fitness landscapes of the Drosophila fly microbiome. H Eble, M Joswig, L Lamberti, WB Ludington. Journal of Mathematical Biology 79 (3), 861-899; 2019
- Microbiome interactions shape host fitness. AL Gould, V Zhang, L Lamberti, EW Jones, B Obadia, N Korasidis, .... Proceedings of the National Academy of Sciences 115 (51), E11951-E11960; 2018
- Microbial quantity impacts Drosophila nutrition, development, and lifespan. ES Keebaugh, R Yamada, B Obadia, WB Ludington, WJ William. iScience 4, 247-259; 2018
- Diet influences host–microbiota associations in Drosophila. B Obadia, ES Keebaugh, R Yamada, WB Ludington, WW Ja. Proceedings of the National Academy of Sciences 115 (20), E4547-E4548; 2018
- Probabilistic invasion underlies natural gut microbiome stability. B Obadia, ZT Güvener, V Zhang, JA Ceja-Navarro, EL Brodie, WW Ja, .... Current Biology 27 (13), 1999-2006. E8; 2017
- Assessing biosynthetic potential of agricultural groundwater through metagenomic sequencing: A diverse anammox community dominates nitrate-rich groundwater. WB Ludington, TD Seher, O Applegate, X Li, JI Kliegman, C Langelier, .... PLoS One 12 (4), e0174930; 2017
- Stable Host Gene Expression in the Gut of Adult Drosophila melanogaster with Different Bacterial Mono-Associations. C Elya, V Zhang, WB Ludington, MB Eisen. PLoS One 11 (11), e0167357; 2016
- Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. MA Koch, GL Reiner, KA Lugo, LSM Kreuk, AG Stanbery, E Ansaldo, .... Cell 165 (4), 827-841; 2016
- A systematic comparison of mathematical models for inherent measurement of ciliary length: how a cell can measure length and volume. WB Ludington, H Ishikawa, YV Serebrenik, A Ritter, RA Hernandez-Lopez, .... Biophysical Journal 108 (6), 1361-1379; 2015
- Avalanche-like behavior in ciliary import. WB Ludington, KA Wemmer, KF Lechtreck, GB Witman, WF Marshall. Proceedings of the National Academy of Sciences 110 (10), 3925-3930; 2013
- Organelle size equalization by a constitutive process. WB Ludington, LZ Shi, Q Zhu, MW Berns, WF Marshall. Current Biology 22 (22), 2173-2179; 2012
- Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. BD Engel, WB Ludington, WF Marshall. Journal of Cell Biology 187 (1), 81-89; 2009
- Automated analysis of intracellular motion using kymographs in 1, 2, and 3 dimensions. WB Ludington, WF Marshall. Three-Dimensional and Multidimensional Microscopy: Image Acquisition and Processing XVI; 2009
- fester, A candidate allorecognition receptor from a primitive chordate. SV Nyholm, E Passegue, WB Ludington, A Voskoboynik, K Mitchel, .... Immunity 25 (1), 163-173; 2006
- MHC-Independent Allorecognition of Invertebrates—A Link between Invertebrate Histocompatibility and Vertebrate Adaptive Immunity? Isolation and Characterization of a Protochordate Histocompatibility Locus. AW De Tomaso, SV Nyholm, KJ Palmeri, KJ Ishizuka, WB Ludington, .... Journal of the American Society of Nephrology 17 (3), 595-599; 2006
- Isolation and characterization of a protochordate histocompatibility locus. AW De Tomaso, SV Nyholm, KJ Palmeri, KJ Ishizuka, WB Ludington, .... Nature 438 (7067), 454-459; 2005
- Genetic variation in Mastocarpus papillatus (Rhodophyta) in central California using amplified fragment length polymorphisms. WB Ludington, KA Callicott, AW Detomaso. Plant Species Biology 19 (2), 107-113; 2004