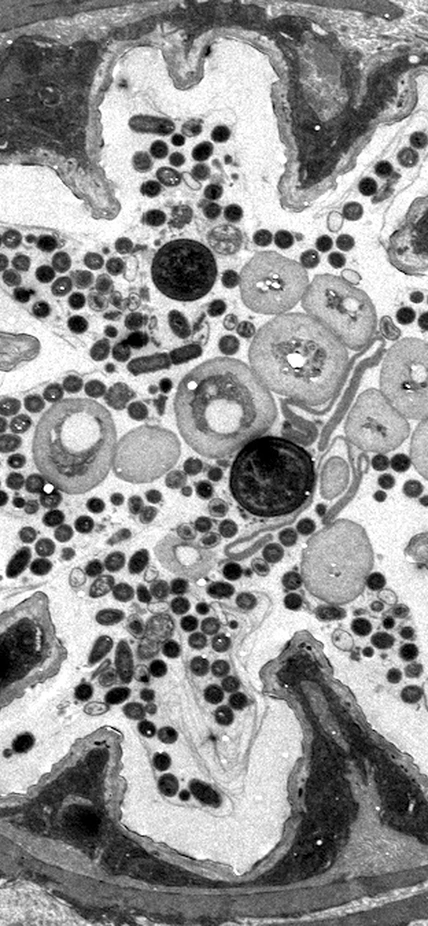
Baltimore, MD—The digestive tract of fruit flies remodels itself to accommodate beneficial microbiome species and maintain long-term stability of the gut environment, according to new research led by Carnegie’s William Ludington and Allan Spradling. Their findings are published in Nature Communications.
The gut microbiome is an ecosystem of hundreds to thousands of microbial species living within the human body. These populations affect our health, fertility, and longevity. But there is still so much to learn about how these microbial species interact with our bodies and with each other.
“Every day, we encounter, and even ingest, a diverse array of bacterial species,” explained Ludington, who has been probing microbiome acquisition and composition for several years at Carnegie. “Despite this, the gut microbiome remains relatively stable over time—a phenomenon that is maintained across many species ranging from mammals to insects.”
He, Spradling, and their collaborators wanted to determine how our guts can maintain such remarkably consistent microbiome compositions. Because the human microbiome is so complex, they studied fruit flies, which are only colonized by a handful of microbial species.
Using sophisticated methods and powerful microscopes, the research team—which included Carnegie’s Ren Dodge, Haolong Zhu, Daniel Martinez, Chenhui Wang, Kevin Aumiller—showed that the fruit fly gut creates physical conditions that selectively promote colonization by certain species.
“The fruit fly gut essentially builds a cozy niche that allows a desirable species of primary colonizers to succeed, fostering a mutually beneficial situation for both the insect and the microbe,” explained Spradling, a longstanding global leader in molecular biology who has developed breakthrough techniques in studying fruit fly genetics.
The researchers found that colonization by a beneficial bacterial strain initiates physical changes in the fruit fly gut that increase the number of binding sites available and produces substances that aid attachment, easing the way for a secondary species to move in.
“Because we’ve discovered this niche in the most powerful model organism for understanding the genetic basis of animal development, it opens up a whole new field of possibilities for understanding the mechanisms by which animals select and control their microbiome,” Ludington concluded.
Their team’s research focused on one particular section of the fruit fly gut that’s transformed into an ideal niche for colonization by two microbial species. Looking ahead, they want to use fly genetics to understand the mechanisms of niche construction and maintenance, as well as search for other potential niches in fruit flies and other animals, including humans.
Other co-authors included Eric Jones and David Sivak of Simon Fraser University, Benjamin Obadia of University of California Berkeley, Andrés Aranda-Díaz and Kerwyn Casey Huang of Stanford University, Zhexian Liu of Johns Hopkins University, Marco Voltolini and Eoin L. Brodie of Lawrence Berkeley National Lab, and Jean Carlson of University of California Santa Barbara.
Acknowledgments
Funding was provided by the Banting and Pacific Institute for the Mathematical Sciences Postdoctoral Fellowships, the Howard Hughes Medical Institute, the Stanford Bio-X Bowes fellowship, the Siebel Scholars program, the Allen Discovery Center at Stanford on Systems Modeling of Infection, the Chan Zuckerberg Biohub, The David and Lucile Packard Foundation and the Institute for Collaborative Biotechnologies through a grant from the US Army Research Office, the Natural Sciences and Engineering Research Council of Canada Discovery Grant, the Canada Research Chairs, the National Institutes of Health, the National Science Foundation, the Carnegie Institution for Science Endowment, and the Carnegie Institution of Canada.