Overview
Grossman has performed research across fields ranging from plant biology, microbiology, marine biology, ecology, genomics, engineering and photosynthesis and initiated large scale algal genomics by leading the Chlamydomonas genome project and serving on the steering committee of the Porphyra umbilicalis genome project.
In 2002 he received the Darbaker Prize (Botanical Society of America) for work on microalgae and in 2009 received the Gilbert Morgan Smith Medal (National Academy of Sciences) for the quality of his publications on marine and freshwater algae. In 2017, he was Chair of the Gordon Research Conference on Photosynthesis and gave the Arnon endowed lecture on photosynthesis in Berkeley in March of 2017. He has given numerous plenary lectures and received a number of fellowships throughout his career, including the Visiting Scientist Fellowship - Department of Life and Environmental Sciences (DiSVA), Università Politecnica delle Marche (UNIVPM) (Italy, 2014), the Lady Davis Fellowship (Israel, 2011) and most recently the Chaire Edmond de Rothschild (to work IBPC in Paris in 2017-2018).
He has been Co-Editor in Chief of Journal of Phycology since 2013 and has served on the editorial boards of many well-respected biological journalism including the Annual Review of Genetics, Plant Physiology, Eukaryotic Cell, Journal of Biological Chemistry, Molecular Plant, and Current Genetics. He has also served on scientific advisory boards for both nonprofit and for profit research institutions and companies including Boyce Thompson Institute, Phoenix Bioinformatics, Exelixis, Martek Biosciences, Solazyme/TerraVia, GEM, Checkerspot and Phycoil.
Research
- Elucidating photosynthesis and nitrogen fixation in the cyanobacteria of the hot springs and how these processes are controlled.
- Understanding the importance of PTOX (plastid terminal oxidase) and the water-to-water cycle in plants, algae and organisms of the oligotrophic oceans (and in other environments).
- Examining the Chlamydomonas genome and using the genomic information to develop insights into gene function (e.g. the GreenCut; informatics analyses that led to identification of various genes/proteins critical for photosynthesis).
- The role of the xanthophyll cycle and LHC family proteins in the dissipation of excess absorbed light energy, a process designated nonphotochemical quenching or NPQ.
- We also identified the first phytochrome-like gene in cyanobacteria that was associated with a function; it is involved in the control of complementary chromatic adaptation (changes in light harvesting pigment composition in response to different light qualities), and characterized the cyanobacterial hli genes, which encode single membrane helix proteins that are likely the progenitors of LHC proteins in plants; the Hli proteins are critical for energy management in cyanobacteria. We (and Don Bryant) were the first to identify phycobiliprotein genes and were among the first to define the ways in which they are regulated. We also identified the regulators of the phosphorus and sulfur deprivation responses in Chlamydomonas… even before they were defined in Arabidopsis (e.g. PSR1, the transcriptional regulator of the phosphorus deprivation responses in Chlamydomonas, which is analogous to PHR1 in Arabidopsis). The more recent directions of the research being done in the laboratory are given below.
The Porphyra genome sequence has been completed and we published a manuscript describing that genome (Brawley et al., 2017). This project incorporated the expertise of many researchers working on marine algae and also led to the publication of two papers on the Porphyra transcriptome (Chan et al., 2012a, 2012b) and another that describes the genetic potential of bacteria that thrive on Porphyra thalli (in this case, three different planctomycetes) (Kim et al., 2016). Again, we expect this information to transform the field and provide novel information about the bacteria that grow on the blades of this macroalga. Many of these bacteria appear to be highly adapted to degrade the complex polysaccharides of the Porphyra cell wall and may be potentially useful in degrading plant cells walls and other polysaccharide-based material. Furthermore, it is also becoming clear that bacteria can impact the development of the macroalgae (e.g. some bacteria produce morphogens).
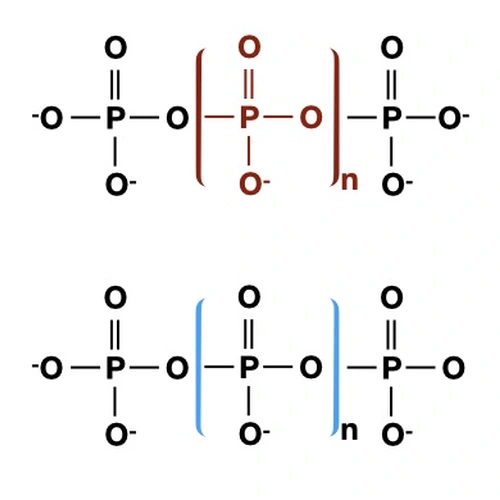
We were the first to identify regulators of the phosphorus and sulfur deprivation responses in Chlamydomonas (Davies et al., 1996, 1999; Wykoff et al., 1999). We are still studying nutrient deprivation responses (Gonzalez-Ballester et al. 2008; 2010; Aksoy et al., 2013; Grossman and Aksoy, 2015), especially with respect to regulatory elements and photosynthetic function (Saroussi et al., 2016; Saroussi et al., 2019). In recent work we also demonstrated that acidocalcisomes are critical for the acclimation of cells to sulfur deprivation (Aksoy et al., 2014), in either a direct or indirect way. The acidocalcisomes may also be a compartment important for storing metals (Sabeeha Merchant’s group). We have begun to methodically examine the various photosynthetic activities (including quenching, cyclic electron flow, linear electron flow, reaction center activity, ATP synthase activity, chlororespiration and the flow of plastid electrons to mitochondrial respiration) under various environmental conditions and in various mutant strains (Saroussi et al., 2016 and 2017), which is expanding our physiological/biochemical understanding of acclimation processes. Recently we made significant inroads into understanding how polyphosphate is a critical component in energy management in Chlamydomonas cells (Sanz-Luque et al., 2020a,b).
Analysis of metabolite trafficking between different subcellular compartments: We have been examining the trafficking of metabolites between different subcellular compartments and especially between the organelles. Mutants have been generated in many chloroplast transporters, including those for triose phosphate and malate/oxaloacetate and the control of those transporters and the phenotypic consequences of null mutations in the transporter genes are being analyzed.
The photoprotective gene: We have been examining the multilayered control of genes encoding proteins involved in the dissipation of excess absorbed light energy in photosynthetic organisms. The controls on these genes include the activity of photoreceptors (both for photosynthetically active light and UV radiation), CO2 levels and the accumulation of reactive oxygen species. This work is leading to a much more thorough understanding the environmental factors that integrate into control these genes and the critical mechanisms associated with photoprotection.
The carbon concentrating mechanism: We are examining the way in which the intracellular structure of the cell rearranges in responses to changes in the CO2 levels experienced by the cells. The rearrangements are extensive and involve changes in the location and arrangement of mitochondria with respect to chloroplasts and potentially the associations between the organelles.
I led the Chlamydomonas genome project, which was performed in collaboration with Dan Rokhsar and Simon Prochnik (as well as several people in the Chlamydomonas community). We were able to give the project a strong biological orientation that led to the development of the GreenCut (Merchant et al., 2007; Karpowicz et al., 2011; Grossman et al., 2019), which has been used to identify novel proteins potentially involved in photosynthesis. The GreenCut is a group of proteins common to green lineage organisms but not present (or present at very low similarity) in heterotrophic organisms. Using a refined analysis, we defined six hundred GreenCut proteins, with nearly half representing proteins of unknown function. This led to the identification of numerous novel proteins associated with photosynthesis, including a number that we have recently characterized; CPLD38 and CPLD49 are involved in assembly/stability of the cytochrome b6f complex and CGL71 is required for assembly of photosystem I (PSI) and appears to protect the PSI complex from oxygen disruption during assembly. A number of manuscripts describing these proteins have been published (Heinnickel et al., 2016; Heinnickel et al., 2013; Wittkopp et al., 2018). Related to this is the work with the Lagarias group on Chlamydomonas hmox mutants, which suggests that a bilin-associated photoreceptor might be involved in photosynthetic control during the transition from low to high/moderate light (Wittkopp, Duanmu et al., 2017). We also developed the concept of the FlagellaCut, which represents a group of proteins involved in the biogenesis/structure/regulation of the flagella. The FlagellaCut analyses allowed us to distinguish between those proteins needed for motility and those needed to satisfy the sensory function of the flagella (Merchant et al., 2007).
The Chlamydomonas genome project required both significant coordination and a strong biological foundation. It changed the ways in which researchers worked with Chlamydomonas (making it an enormously powerful model organism) and led to new work in which Martin Jonikas and I (but mostly Martin) developed an indexed mutant library of Chlamydomonas, for which the genome insertion sites are mapped; this library covers over 70% of the Chlamydomonas genes (having at least one mutant allele). The work on the initial library and a second paper describing an expanded library and its uses were recently published (Li et al.,2016; Li et al., 2019). Global analyses of the library for evaluating gene function is being reviewed at Nature Genetics (Fauser et al., 2021). Mutants in the library are available from the Chlamydomonas Resource Center with thousands of mutants that have already been ordered (and numerous ones published on).
We are using the Aiptasia-Symbiodiniaceae model system to explore interactions between the animal (Aiptasia, which is a sea anemone/cnidarian; corals are also cnidarians) and its algal endosymbiont. We have isolated many axenic Symbiodiniaceae strains (Xiang et al., 2013), developed a strong collaboration with John Pringle to exploit the Aiptasia system (Bieri et al., 2016) and showed that the bleaching of the holosymbiont is provoked by elevated temperature in the light or dark (Tolleter et al., 2013). We have also performed a lot of work with the cultured Symbiodinaceae alga Breviolum minutum (which can infect Aiptasia) and showed that it can grow rapidly under heterotrophic conditions (on glucose in the dark) and that in the light on glucose it bleaches and loses its photosynthetic apparatus (Xiang et al., 2013; Xiang et al., 2015; Xiang et al., 2016). Our work on the coral association has extended to collaborations with Simon Davy’s group in New Zealand (Oakley et al., 2016; Oakley et al., 2015 and 2017; Mathews et al., 2015, 2107, 2020; Sproles et al., 2016 and 2018; Gorman et al., 2020; Sproles et al., 2020a.b). We also have evidence showing that after Symbiodinium SSB01 enters and fully populates the Aiptasia host, it becomes limited for nitrogen within the host, and that the nitrogen-carbon exchange likely plays a strong role in controlling the symbiotic association. Work is currently underway to understand the role of photosynthesis and nitrogen in the infection/population of the host, elucidation of the ways in which sugars change the physiology of the symbiont and host and alter their interactions, elucidation of acclimation responses (light levels and temperature through RNA-seq and TEM analyses) and studies associated with the events involved in degradation of the coral-algal association (e.g. elevated temperature). We are also attempting to transform Breviolum minutum and other algae of the Symbiodiniaceae and generate gene knockout lines. Furthermore, we recently participating in analyzing the extremely tightly packed nuclear genome structure of Breviolum minutum, which was recently accepted for publication in Nature Genetics.
The establishment of plastids in photosynthetic eukaryotes has been attributed to a single primary endosymbiotic event that occurred ~1.6 BYA in which a cyanobacterium was engulfed and retained by a protist. Currently, we know little about early events in plastid evolution. Paulinella chromatophora (photosynthetic amoeba) represents a unique model to explore plastid evolution because it contains a cyanobacterium-derived photosynthetic organelle termed “chromatophore” that originated ~100 MYA. The chromatophore has a genome of ~1/3 the size of a typical unicellular cyanobacterial genome; this genome is likely experiencing genome reduction. Several genes from the ancestral cyanobacterium were transferred to the ‘host (protist)’ nuclear genome in a process called endosymbiotic gene transfer (EGT). At least some of the EGT proteins are routed to chromatophores after being translated outside of the organelle on cytoplasmic ribosomes (the chromatophore is therefore a canonical organelle). This was demonstrated for two cytoplasmically synthesized subunits associated with PSI (Nowack et al., 2012). A number of EGT-derived genes encode proteins integral to photosynthesis and photoprotection, with the hli gene family likely expanding in the host nuclear genome (Zhang et al., 2016). Additionally, both genomic and transcriptomic sequences were used to analyze integration of functionalities encoded on the chromatophore and protist nuclear genomes. Metabolic pathway reconstruction showed that the P. chromatophora nuclear gene inventory appears to complement gene loss on the chromatophore genome. Surprisingly, this is not predominantly achieved through EGT from the nascent organelle but is mostly achieved by retailoring of host genes to compensate for loss of chromatophore genes and also through horizontal gene transfer (HGT) from diverse bacterial sources (Nowack et al., 2016). This latter finding may reflect the phagotrophic life-style associated with the protist as it was becoming a photoautotroph (the protist was still consuming bacteria when the cyanobacterium first became an intracellular resident). More recent work shows that some genes derived by EGT evolved gene families that expanded through retrotransposition and are critical for the new life style experienced by the photosynthetic amoeba (Gabr et al., 2020; Stephens et al., 2021; Calatrava et al., 2021).
Press Releases
CV
Postdoc, 1978 - 1982: Rockefeller University, Uptake of Polypeptides Into Chloroplasts
Ph.D., 1978: Indiana University, Characterization of Photosynthetic Mutants in Chlamydomonas reinhardtii
B.S. (With Honors), 1973: Brooklyn College, Biology
- 2021 - Editor of the Chlamydomonas Sourcebook
- 2021 - Search Committee for Life Science Director – Carnegie Institution
- 2021 - Search Committee – Senior Staff Scientist – Carnegie Institution
- 2021 - Search Committee – Staff Associate – Carnegie Institution
- 2021 -Co-editor-in Chief, Journal of Phycology (from 2012)
- 2021 - Caltech Biosafety Committee
- 2021 - Editorial Board, Current Genetics (from 2000)
- 2021 - Building Committee, Life Science Building in Pasadena for Carnegie Institution
- 2021 - External Jury Member for Florence Mus Habilitation à Diriger des Recherches
- 2021 - Boyce Thompson Institute – Scientific Advisory Board (from 2014)
- 2019 - NSF Panel for Science and Technology Center
- 2019 - DOE-BES Panel
- 2018 - Organizer of the International Conference on the Genetics and Molecular Biology of Chlamydomonas
- 2018 - Organizing Committee, International Conference on Microbial Photosynthesis
- 2017 - Edmond de Rothschild Chair Fellowship to work at Institut de Biologie Physico-Chimique, Paris
- 2017 - Arnon Endowed Lecture – Berkeley
- 2017 - Chair of the Gordon Conference on Photosynthesis
- 2016 - Review Panel for the Integrated Microbial Diversity Program/Canadian Institute for Advanced Research
- 2015 - Review Panel for Genomics and Synthetic Biology Program, NYU, Abu Dhabi
- 2015 - Co-Chair of the Gordon Conference on Photosynthesis
- 2014 - Visiting Scientist Fellowship - Università Politecnica delle Marche
- 2017 - Steering Committee, University of Texas Culture Collection (from 2011)
- 2014 - Review of Science and Technology Center, University of Nebraska
- 2013 - Organizer of Western Photosynthesis Conference
- 2013 - Participant in17th International Course of School of Pure & Applied Biophysics, Venice
- 2013 - External examiner on thesis defense of Laura Houille, IBPC, Paris, France
- 2013 - Member of EU Sunbiopath Evaluation committee
- 2013 - Panel Member to Review Science and Technology Center, University of Nebraska
- 2013 - Review of Science and Technology Centers for NSF
- 2012 - Co-organizer of Western Photosynthesis Conference
- 2012 - Editorial Board, Journal of Phycology (from 1997)
- 2011 - Member of DOE Evaluation Committee, Plant Research Laboratory
- 2011 - Recipient of Lady Davis Fellowship (Israel)
- 2009 - Steering committee, Joint Genome Institute
- 2009 - Recipient Gilbert Morgan Smith Medal (National Academy of Sciences)
- 2009 - Editorial Board Annual Review of Genetics (from 2005)
- 2008 - National Resources Defense Council Algal Biofuels Advisory Committee
- 2007 - Editorial Board, Eukaryotic Cell
- 2008 - Editorial Board, Molecular Plant
- 2004 - Geographical representative for the International Society of Photosynthesis Research
- 2004 - Editorial Board, Plant and Cell Physiology (from 2000)
- 2000- Scientific Advisory Board for the Wallenberg Consortium North (from 2000)
- 2002 - Darbaker Prize for work on microalgae (Botanical Society of America)
- 2000 - Advisory Committee, Arizona State University Consortium for NSF funded Biotechnology Cente
- 2000 - Organizer of Symposium ‘The Dynamics and Evolution of Light Harvesting Complexes
- 1999 - Co-organizer of Symposium to Honor the retirement of Olle Bjórkman
- 1998 - Organizer of US-Japan Binational Meeting (Asilomar, CA)
- 1998 - Editorial Board, Journal of Biological Chemistry (from 1996)
- 1996 - Guest on the Editorial Board of the Annual Review of Genetics
- 1995 - Plenary Lecture - Japan Society of Plant Physiologists
- 1995 - Coordinator for the Organization of the Plant Biology Retreat, Stanford University
- 1994 - Member of AIBS panel to evaluate joint projects in Plant Biology
- 1994 - Editor-Seminars in Cell Biology "Light regulation in photosynthetic organisms"
- 1994 - Organizer of the Plant Biology Retreat, Stanford University
- 1993 - Guest Editor of the Annual Review of Genetics
- 1993 - Organizer: Cyanobacterial Workshop at Asilomar, CA
- 1993 - Guest lecturer in Marine Molecular Phycology Course at Friday Harbor Marine Station
- 1992 - Recipient-Nehru University, Dept of Biotechnology Fellowship
1992 - Organizer of the Carnegie Institution Plant Biology Seminar Series
1991 - Member Photosynthesis Panel (USDA)
1991 - Co-organizer, conference on tetrapyrroles (with Paul Castelfranco)
1990 - Editorial Board Journal of Plant Growth Regulation
1989 - NSF Panel - Postdoctoral Fellowship Awards
1988 - Organizer of the Carnegie Seminar Series
1988 - Co-Organizer of C. Stacy French Symposium on Photosynthesis.
1988 - NSF Panel - Postdoctoral Fellowship Awards
1987 - Program Committee “Molecular Biololgy of Cyanobacteria Workshop, St. Louis, MO
1987 - Photosynthesis Panel (USDA)
1987 - Organizer of the Carnegie Seminar Series
1987-93 - Editorial Board, Plant Physiology
1985 - Guest Editor Annu Rev Plant Physiology
1985 - Plant Molecular Biology Panel (USDA)
1984 - NIH Postdoctoral Fellowship Panel
1979-81 - Postdoctoral Fellowship Panel (NIH)
1977 - Floyd Fellowship
1974-77 - National Science Foundation Predoctoral Fellowship
1972 - L. Whorley Award in Biology
1968-72 - New York State Regents Scholarship Phi Beta Kappa
- Chief of Genetics, Solazyme (2007-2015) – Algal Biotechnology
- Martek Biosciences Corporation (1993-2000)
- Exelixis Pharmaceuticals (1995-2001)
- Phoenix Bioinformatics (2013-2018; 2020-Current)
- Checkerspot (2016-Current)
- Phycoil (2016-Current)
- GEM Health (2020-Current)