A new Venture Grant has been awarded to the Geophysical Laboratory’s Dionysis Foustoukos and Sue Rhee of the Department of Plant Biology, with colleague Costantino Vetriani of Rutgers University for their project Deciphering Life Functions in Extreme Environments.
Carnegie Science Venture Grants ignore conventional boundaries and bring together cross-disciplinary researchers with fresh eyes to explore different questions. Each grant provides $100,000 support for two years with the hope for surprising outcomes. The grants are generously supported, in part, by trustee Michael Wilson and his wife Jane and by the Ambrose Monell Foundation.
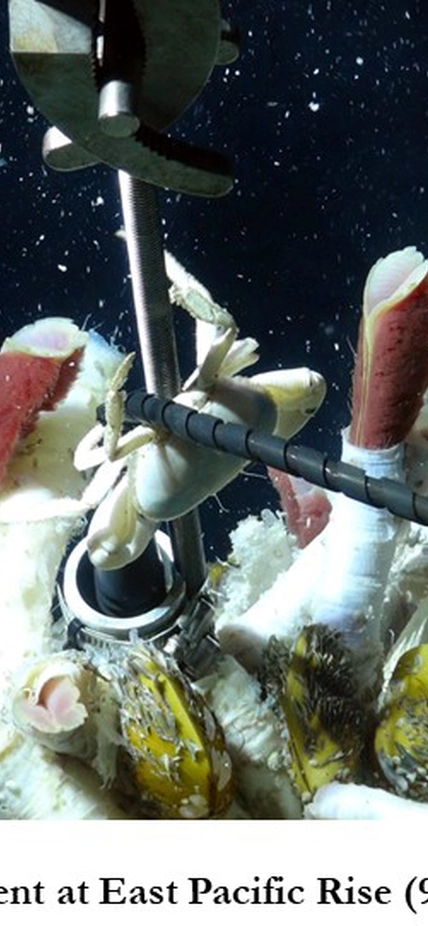
Deep sea hydrothermal vents support a remarkable diversity of life, without sunlight, limited nutrients, and in extreme pressure and temperature conditions. Without photosynthesis, life there has evolved to convert rock chemistry into usable energy. Despite the fact that these microorganisms are major contributors to global biogeochemical cycles, very little is known about their function and physiological responses to the extreme conditions of deep-sea habitats.
Foustoukos, Rhee, and Vetriani have teamed up to integrate microbial physiology, genomics, and metabolic network modeling with high pressure and temperature experimentation to understand gene regulation in response to changing environmental conditions. Their objective is to unravel how microorganisms interact with each other and the environment. They will use a high-pressure adapted (piezophilic) chemolithoautotrophic bacterium that Foustoukos and Vetriani recently isolated from an active deep-sea vent at the East Pacific Rise. This is the only autotrophic organism that has been characterized among the piezophilic organisms isolated to date.
The scientists will look at the adaptation mechanisms of the microorganisms’ metabolism in response to different pressures up to 680 times atmospheric pressure and temperatures up to 175º F (80 º C) in different nutrients using a novel bioreactor used by Foustoukos and Vetriani. Experimental results will be compared to genomic studies and genome databases from the Rhee lab to reconstruct the metabolic network models of piezophilic microorganisms.
The team hopes to better understand how environmental factors and physiology of these organisms shape the evolution of the deep biosphere.