Bold Initiatives
Can We Open the Door to a Reliable Life Detection Method?

Earth and Planets Laboratory Staff Scientist Anat Shahar is the lead investigator on an interdisciplinary, multi-institution research team that in the spring of 2021 was awarded nearly $1.5 million from the Alfred P. Sloan Foundation to understand the chemical makeup of our galaxy’s most common planets with a goal of developing a framework for detecting chemical signatures of life on distant worlds.
Since the first planet orbiting a Sun-like star beyond our Solar System was discovered in the mid-1990s, astronomers have revealed that the Milky Way is teeming with planets. To search for evidence of life on these thousands of other worlds, researchers are on the hunt for chemical markers that could only be produced by biological processes. This means they must be able to rule out every possible non-biological origin for these chemical signatures.

To understand the non-biological processes that determined a planet’s atmosphere, you have to understand its formation and evolution. Carnegie’s longstanding leadership in the geosciences, planetary science, and astronomy make this the logical home for such an ambitious undertaking. And the collegial community at the Earth and Planets Laboratory campus in Washington, D.C., fosters collaboration across a suite of relevant physical sciences.
For decades, our researchers have developed expertise in using both laboratory experiments and model development to reveal the conditions found in the interiors of our own and other planets. Furthermore, our astronomers have unparalleled access to the world-class telescopes at our Las Campanas Observatory in Chile.
Shahar’s Atmospheric, Empirical, Theoretical, and Experimental Research (AEThER) project aims to determine the abiotic atmospheric composition of sub-Neptunes, as well as other, smaller rocky bodies. Although our own Solar System doesn’t have any sub-Nepture or super-Earth sized planets, they are the most common type of exoplanet discovered so far.
A planet’s atmosphere is influenced by its accretion from the dust and gas that surrounds its host stars in its youth, and by the differentiation of this material into distinct layers. It is later additionally modified by geologic activity on the surface, including volcanism and tectonics.
Shahar and her team—including colleagues and collaborators from Carnegie, Howard University, University of Washington, the Observatoire de Paris, the Harvard & Smithsonian Center for Astrophysics, University of Maryland, University of Rochester, Oxford University, and UCLA—aim to reveal the intricate connections between a planet’s interior, surface, and atmosphere in order to eliminate every possible “false positive” biosignature. They believe this will open the door to a reliable method of life detection.

The next generation of ground- and space-based telescopes, including the Giant Magellan Telescope under construction at Las Campanas, will enable astronomers to determine the compositions of gas giant exoplanet atmospheres with never-before-seen detail. But to truly develop a foolproof method for detecting biosignatures, more than one area of expertise is needed, which is why AEThER will deploy a multi-disciplinary approach, including observations, experiments, and modeling.
Just as a diversity of discipline and methodology is necessary to tackle such a large challenge, so is a diversity of experience. Another component of the Sloan grant will fund training and mentoring undergraduate and graduate students with the hope of increasing representation among the next generation of Earth and planetary scientists. Bringing additional perspectives to the table will open up the possibility of new ideas and creative approaches to scientific problem-solving.
What Makes Plant Cells Special?
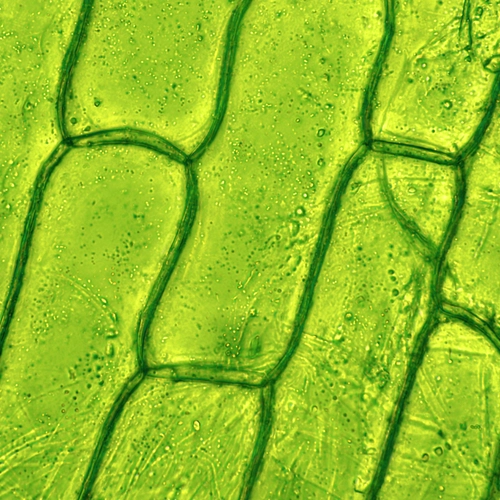
The world’s population is growing, and global climate change will reshape our maps—shifting locations where human settlements can sustainably exist and thrive. The World Health Organization predicts that malnutrition, infectious disease, and extreme heat caused by climate change will cause about a quarter of a million deaths each year between 2030 and 2050. Related economic losses are projected to be several hundred billion dollars a year in the U.S. alone by 2090. And we are losing species at the highest rate we have seen since the dinosaurs went extinct; a phenomenon that scientists have coined “the sixth extinction.”
Plant science can help us understand and mitigate the coming challenges, including fighting hunger, promoting renewable energy, and sequestering carbon pollution from the atmosphere. But in order to meet the moment, the scientific enterprise must prepare to leap ahead in its understanding of how plant cells function and respond to their environmental conditions. And to successfully advance plant science, the scientific community must foster the next generation of researchers and to ensure that a premium is placed on inclusivity and diversity in laying this foundation for the future.

Courtesy Carnegie Institution for Science.
In 2019, Carnegie’s Sue Rhee and David Ehrhardt, along with NYU’s Kenneth Birnbaum launched the Plant Cell Atlas initiative and called on the research community to increase its investment in plant sciences with the goal of significantly advancing available knowledge of the links between plant cell structure, biochemistry, and physiology. They argued that the iron is hot to reimagine plant science as critical not only to basic scientific research, agriculture, and environmental stewardship, but to energy, healthcare, and manufacturing—a major economic and cultural force.
Due to recent innovations in data science, artificial intelligence, biosensors, systems biology, gene editing, precision breeding, and microbiome research, Rhee, Ehrhardt, and Birnbaum felt that the field is well positioned to drive major breakthroughs. They added that the endeavor would require plant scientists to team up with biologists from biomedical fields, as well as experts in nanotechnology, imaging, and scientific computing.

Through a series of three well attended and highly collaborative kickoff workshops, more than 400 experts in a variety of fields discussed a roadmap for spring-boarding our understanding of plant cells into a new era. Based on these extensive conversations, the members of this multi-institution, interdisciplinary collaboration identified a set of goals for significantly advancing our understanding of how plants accomplish their amazing cellular, physiological, and biochemical feats.
Some of these aims are technological, involving the creation of new tools and techniques. Others focus on information sharing. But fundamentally the project wants to identify and map every type of plant cell and the location and organization of the various biochemical products found inside them.
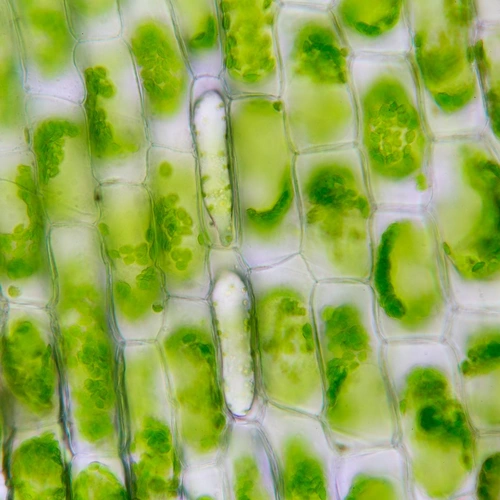
Unlike animal cells, plant cells can convert the Sun’s energy into chemical energy—sugars and fats—using a biochemical process called photosynthesis. This forms the baseline of our entire food supply, both directly through the consumption of fruits, vegetables, legumes, grains, and other plant products and, for meat eaters, indirectly through consumption of plant-eating livestock.
Photosynthesis has also shaped our planet’s atmosphere. Its evolution made Earth oxygen rich, allowing life as we know it to arise. Furthermore, its reactions require carbon dioxide, which means that plants take up and sequester about a quarter of the carbon pollution released by human activity.
Plants are also remarkable in that many of them can withstand extreme environments that would kill most animals. For example, there are desert plants that thrive at temperatures as high as 116 degrees Fahrenheit (47 degrees Celsius), which is 12 degrees higher than the maximum temperature on a standard hot tub. How plants evolved to become resilient to such harsh environments and what tools they use to perform such astonishing feat remain a mystery.

Art and science exhibit featuring agricultural transformer George Washington Carver and pollination prodigy Edmond Albius at Bowie State University.
Another goal of the Plant Cell Atlas is to nurture the future leaders of plant biology. Seventy percent of workshop participants were early career scientists. In addition to reflecting a diversity of scientific disciplines and a bottom-up ethos, the community is committed to teamwork, mutual respect, active listening, and psychological safety.
As part of its efforts to make a transformative impact on diversity in plant science, the Plant Cell Atlas community developed a science and art exhibit at three Historically Black Colleges and Universities—Howard University, Morgan State University, and Bowie State University—highlighting the work of four Black plant scientists through history. Each display was accompanied by suggested classroom activities.
Far too often, the Plant Cell Atlas organizers asserted, BIPOC plant scientists have had their work overlooked or their roles diminished in the public discourse, which makes others feel unwelcomed. Plant science is too crucial to humankind’s survival to leave anyone out of the conversation. These efforts recognize that because science is all about creativity and problem solving, having as many voices as possible at the table will result in a wider array of ideas and approaches and help everyone surmount the coming environmental challenges.
How Microbial Communities Shape Our Lives

William Ludington, courtesy Carnegie Institution for Science.
Since William Ludington’s arrived at Carnegie’s Department of Embryology in 2018, he has been aggressively pursuing breakthroughs in microbiome research—deploying a multitude of genetic, physiological, and mathematical approaches to understand the community ecology of our gut microbiome and how it affects our health, fertility, and even longevity.
The human microbiome is an ecosystem of hundreds to thousands of microbial species living within our gastrointestinal systems. The tremendous species diversity of these communities makes it challenging to elucidate how its various parts add up to a greater whole. In other words, to what extent do individual species influence our health and physiology, and to what degree are these impacts determined by interactions between the species present in our microbiomes?
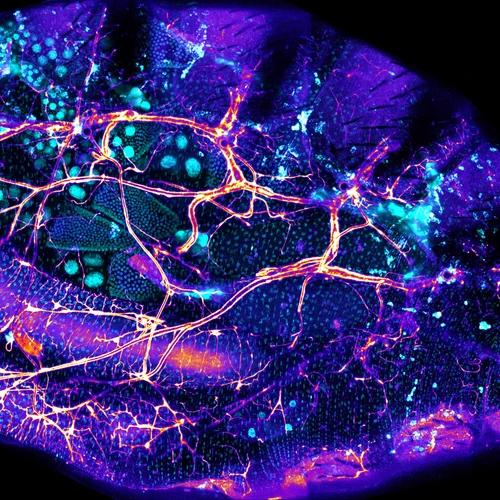
The microbiome as a is like a classic Hollywood musical number starring bacteria and being filmed inside your gastrointestinal system—each dancer is doing something interesting, but it’s only when the camera pulls back to see the whole company from overhead that you understand the magnitude of the piece. Ludington’s lab uses various techniques and approaches to probe these myriad relationships and attempt to reveal how the microbiome functions as a collective unit.
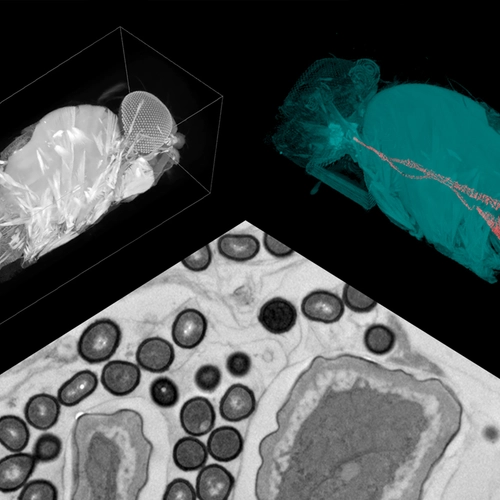
In particular, the Ludington lab investigates the dynamics of microbial interactions using the fruit fly, Drosophila melanogaster. The fruit fly gut carries far fewer microbial species. Using these insects to understand species-species interactions in the gut microbiome could eventually drive improvements in precision medicine and guide the development of regenerative therapies.

Ludington Lab manager Daniel Martinez studies a micro-CT scan of a fly gut. Photograph by Navid Marvi, courtesy of the Carnegie Institution for Science.
In 2021, Ludington’s quest to answer big questions about our microbiome was recognized with two funding windfalls that will propel his work to a new level of excellence. He was named a recipient of nearly $1 million over three years from the National Science Foundation and selected as one of 14 researchers to receive $55,000 from the Research Corporation for Science Advancement for its inaugural Scialog: Microbiome, Neurobiology, and Disease initiative. Short for science + dialog the awards were sponsored by RCSA, The Paul G. Allen Frontiers Group, and the Frederick Gardner Cottrell Foundation.
His NSF-funded project, jointly led with Stanford University’s Kerwyn Huang, aims to combine cellular physiology and ecological theory to understand the link between the actions of single cells and their success in the gut microbial ecosystem. Knowing how an individual cell’s response to its environmental surroundings affects its evolutionary fitness in the microbial community would be a major advance in the field. It could enable microbiome engineering to improve human health, increase agricultural yields, protect freshwater resources, and more.

Understanding the role evolutionary principles play in microbiome communities could also help explain the microbiome’s profound impact on neurological health and function. Age-related loss of microbiome species diversity is associated with neurological disorders in fish, mice, and humans. Ludington and UCLA’s Nandita Garud hypothesize that the mechanism behind this neurodegeneration is evolutionary, rather than ecological. Their Scialog project could cause in a fundamental shift in the approach to microbiome research by investigating relationships between genetic changes in the microbiome and age-related frailty.
RCSA prides itself on funding early-stage, high-risk, potentially high-reward science. RCSA is willing to bet on new and innovative ideas and accepts that up to 50 percent of projects may fail. Using a proprietary algorithm designed to spur collaboration, the Scialog grouped together chemists, physicists, biologists, and neurophysiologists to investigate linkages between the microbiome and neurodegeneration. Scientists were then invited to submit project proposals with others whom they met at the conference.
“Will’s work to understand how an individual cell responds to its environment and affects its evolutionary fitness could dramatically change how we think about the microbiome,” said Andrew Feig, Senior Program Director for RCSA. “We are excited by the possibilities of this innovative partnership.”