Modern sequencing platforms, such as the Illumina HiSeq 2000, generate only short, ordered sequences, usually 100 consecutive bases—adenine, guanine, cytosine, and thymine—in each reaction. But by doing this on billions of molecules in parallel, these sequencers generate between 100,000 and 1 million times more data than previous methods. That’s where Frederick Tan fits in. Departmental scientists cope with this data avalanche by combining their biological insights with complex computer algorithms and statistical methods. Tan manages the genomics and bioinformatics facilities and guides others on their use.
Tan shares his knowledge by leading workshops, group-study sessions, and teaching courses to familiarize Carnegie and Johns Hopkins researchers with bioinformatics approaches. He started a study group called Data Wranglers Anonymous where people learn to handle large numbers of data files, conduct exploratory data analyses, and write programs to extract useful information. Tan also created a series called Nitty Gritty Workflows to discuss sequence analysis pipelines. Speakers describe software programs they used and why they chose specific parameters—important for troubleshooting and establishing best practices. He helped Embryology host the fourth annual Practical Genomics Workshop and has ambitious plans for future training.
Current Topics
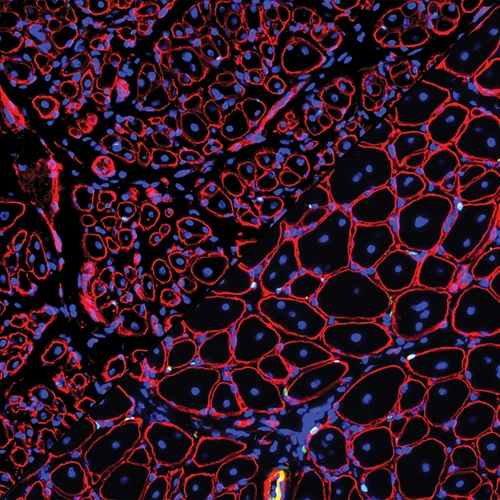
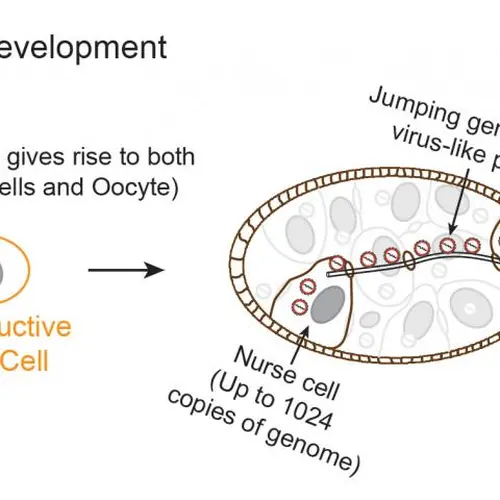
Such factors determine how often non-allelic repetitive DNA recombines to produce chromosomal aberrations associated with cancer and hereditary diseases. To address this question, I am developing genome-wide tools to determine what factors are most important in governing recombination between naturally occurring repeats. In addition to spontaneous recombination events, recombination machinery can also be harnessed as a powerful tool to modify genomes precisely. I am using genetic screens to identify rate-limiting steps during genome modification.