Stanford, CA— You’ve probably seen news stories about the highly lauded, much-discussed genome editing system CRISPR/Cas9. But did you know the system was actually derived from bacteria, which use it to fight off foreign invaders such as viruses? It allows many bacteria to snip and store segments of DNA from an invading virus, which they can then use to “remember” and destroy DNA from similar invaders if they are encountered again. Recent work from a team of researchers including Carnegie’s Devaki Bhaya demonstrates that some bacteria also use the CRISPR/Cas system to snip and recognize segments of RNA, not just DNA. It was published by Science.
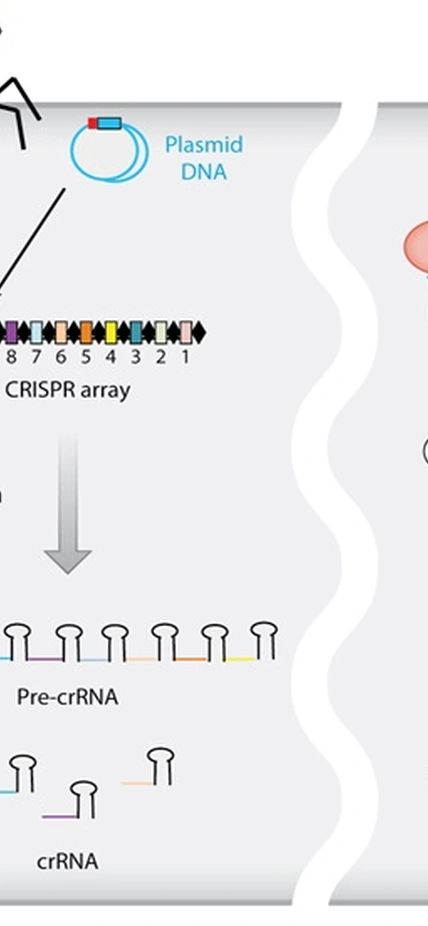
A quick primer on CRISPR/Cas systems: A few decades ago, biologists noticed that many bacterial genomes contain small repeated segments of DNA that were interspersed with segments of “mystery” DNA. They christened them CRISPR for “clustered regularly interspaced short palindromic repeats.” Years later they figured out that the unique segments of DNA often came from invading viruses. The bacteria’s complex of Cas enzymes snips and incorporates virus DNA into these CRISPR regions. This arsenal of snipped segments of viral DNA helps the bacterium to recognize identical invading DNA, after which the Cas protein machinery moves in to destroy it. Very recently, scientists figured out that this system could be repurposed into precision-guided gene-editing tool that is faster, cheaper, and more accurate than any previous options.
There are several types of CRISPR/Cas systems, and in some of them, there is a novel fusion protein in which a Cas enzyme is fused with a protein called a reverse transcriptase. Reverse transcriptase is able to read a strand of RNA and use it to construct a complementary strand of DNA. The existence of this novel protein made the research team wonder if it could be used to confer immunity against an RNA-based invader.
Working with the marine bacterium Marinomonas mediterranea, the team was able to demonstrate that the reverse transcriptase-fused to a Cas protein can, indeed, use RNA and integrate it into CRISPR regions.
Bhaya explained: “The team has demonstrated that this biochemical process can occur in the lab, and based on this information, the CRISPR/Cas system may confer immunity against RNA-based invaders out there in the wild.”
The team was comprised of Stanford University’s Sukrit Silas (graduate student and lead author), Georg Mohr, and Andrew Fire (a former staff member at Carnegie’s Embryology Department); David Sidote, Laura Markham, and Alan Lambowitz of the University of Texas at Austin; and Antonio Sanchez-Amat at the Universidad de Murcia in Spain.
“It is gratifying to see how much we can learn from the extraordinary protein diversity that exists in the microbial and viral world, especially when it is combined with rigorous biochemistry,” said Bhaya. She added “I think our colleague, Karl Deisseroth, at Stanford University, summed it up best when he said: ‘We need to be supporting the people who are fascinated by pond scum and other obscure topics if we are to eventually treat depression, autism, Parkinson’s disease and a host of other complex diseases.’”
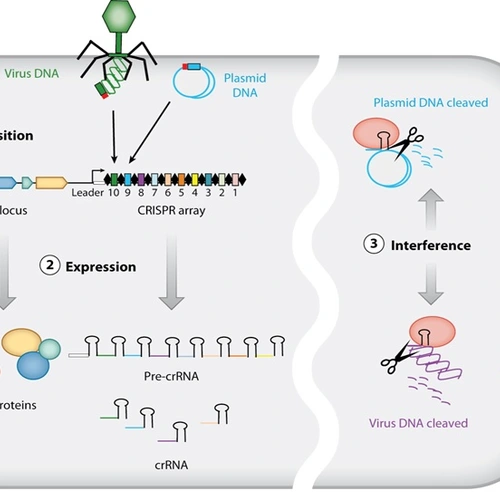
Caption: An illustration of how the CRISPR/Cas system works, courtesy of Devaki Bhaya, Michelle Davison, and Rodolphe Barrangou originally published in Annual Review of Genetics.
__________________
This work was funded by NIH, an HHMI International Student Research Fellowship, the Stanford Graduate Fellowship, and the Welch Foundation.