A First: Connecting Genomics and Ecological Modeling
Human-induced climate change and exploitation increased species’ extinction rate over 1,000-fold in the 20th century—50% suffered local extinctions. However, researchers have not been able to predict which species can and cannot adapt. Until now, ecological adaptation models have not included genomic information. Carnegie’s Moises Exposito-Alonso’s team investigated how adaptation is molecularly encoded in DNA and developed predictive models of future extinction risk.
This method has roots in the legendary 1941 experiments of Carnegie’s Jens Clausen, David Keck, and William Hiesey, who planted seeds of common species of different California plant populations. When they planted seeds of the same species in environments close to seed collection points, they grew best, indicating they genetically adapted despite looking almost identical. Without today’s technology, the team could not pinpoint specific adaptive mutations. Expósito-Alonso and colleagues used this concept to understand species’ susceptibility to changing climate conditions using the model plant in genetics, Arabidopsis thaliana, which grows naturally in different climates around the world.

From left to right, Carnegie’s Jens Clausen, David Keck, and William Hiesey worked in plant ecology and pioneered the study of local adaptation in the 1940s. Image courtesy Carnegie Institution for Science
They conducted experiments—similar to those of Clausen, Keck, and Hiesey— in Germany and Spain, simulating expected precipitation changes in Europe. They planted about half a million seeds from 500 European locations and studied their growth and survival. They found specific genetic differences in the plants’ DNA. Remarkably, similarly looking plants, grown in the same soil with the same water, had different outcomes: some shriveled and died, while others remained green and flowered. The team developed algorithms to sort through 10 million DNA changes and identified several thousand that best explained the differences.
With these results, they developed the first ecological risk model of population maladaptation under climate change that accounts for genome-wide mutations of each natural population. The prediction map showed how lower precipitation trends in Europe will decrease adaptation and increase risk of local extinction of Central European populations, which have not experienced past droughts. These populations differ from Mediterranean Arabidopsis, which have mutations that could increase survival under future heat and drought conditions.
The team now wants to test these climate adaptation models with a globally distributed experiment and a consortium network called Genomics of rapid Evolution in Novel Environments (GrENE-net). In the long run, these models will be applied to keystone ecosystem species to understand climate-change resilience.
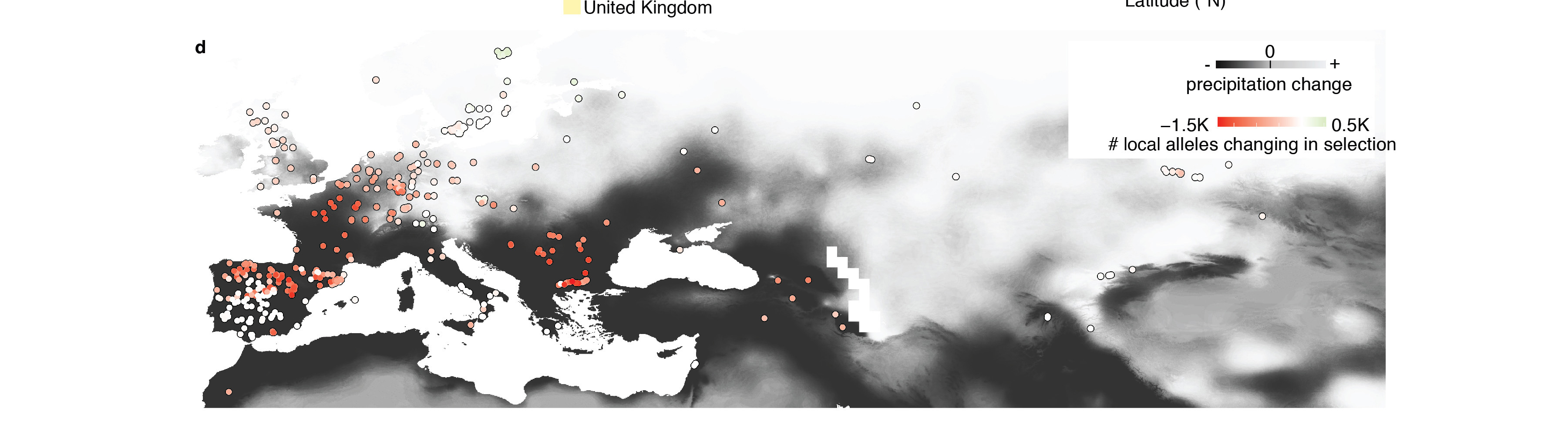
This map is one product of the new model showing adaptability. Populations of the model plant Arabidopsis in central Europe had a variant, which is good for a wet climate but not for drought. White indicates less risk to decline by 2050, while red indicates more risk.

Images courtesy of Moises Exposito-Alonso.

