Research Interests
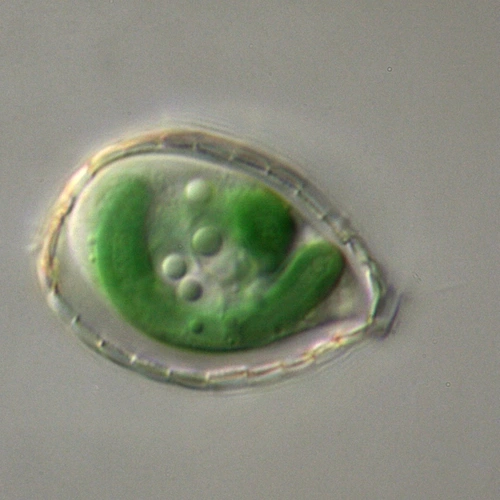
Research in our lab is driven by an interest in understanding how photosynthetic microorganisms perceive and evolve in response to environmental fluctuations of light, nutrients and phage attack. We focus on an ancient and diverse group of microbes called cyanobacteria. They are
abundant, globally relevant and have been used to probe important processes such as photosynthesis, symbioses and circadian rhythms. Recently their environmental impact through toxic blooms and possible use as green chassis for high value products have gained attention.
I believe that collaboration rather than competition is a healthy research environment to strive for. Consequently, we work with several groups at Stanford and elsewhere. We use a toolbox that includes biochemistry, microscopy, genetics, bioinformatic pipelines, synthetic biology, statistical
methods. Several of us enjoy teaching, communicating science to a broad audience and feel this is an important activity.
We work both with both model cyanobacteria and with novel cyanobacterial isolates from naturally occurring communities. The lab has a current focus on querying a comprehensive 'omics dataset' acquired from the microbial mats in Yellowstone National Park, to understand resource-sharing
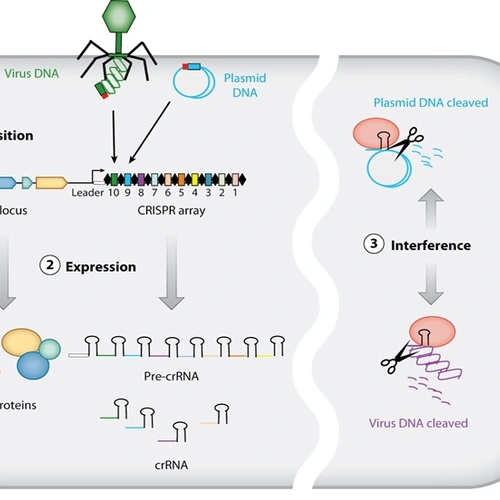
fluctuations and phages in extreme environments. Projects in the lab include understanding phototaxis; the behavior of co-cultured organisms, developing genetic tools for undomesticated microbes and a long-term goal to build synthetic microbial communities.
If this sounds interesting, feel free to contact me, visit our campus, do a lab rotation or a research project.
Collaborators
- Andrew Fire (Sukrit Silas), Dept. of Pathology &Genetics, School of Medicine, Stanford University.
- Arthur Grossman, Carnegie Institution for Science (funded by NSF grants)
- Brian Yu, CZ Biohub, Stanford University (JGI- Large scale CSP grant)
- Daniel Fisher (Mike Rosen, Gabriel Birzu), Dept. of Applied Physics, Stanford University
- Dave Ward, Fred Cohan, John Heidelberg (funded via Emerging Frontiers Grant)
- John Golbeck, Chris Voigt, Susan Rosser, Bill Rutherford
- KC Huang (Rose Chau, Tristan Ursell), Dept. of Bioengineering, Stanford University (funded by Stanford BioX grant)
- Mihai Pop, Jackie Meisel, Dept. Computer Sciences, University of Maryland
- Seppe Kuehn, Alison Smith, Chris Howe (funded by NSF-BBSRC grant)
- Todd Treangen, Santiago Segarra, and Luay Nakhleh, Dept. of Computer Sciences, Rice University (funded by NSF grant)